Genome Editing Used to Correct Mitochondrial Mutations
By LabMedica International staff writers
Posted on 08 Oct 2018
A recently developed mouse model, which mimics a cardiac mitochondrial disease, was used to demonstrate the potential use of genomic engineering to treat the disorder by eliminating the mutation that causes it.Posted on 08 Oct 2018
Mitochondrial diseases are, as the name implies, a group of disorders caused by dysfunctional mitochondria. About 15% of the time these diseases are caused by mutations in the mitochondrial DNA (mtDNA) that affect mitochondrial function. Mitochondrial diseases take on unique characteristics both because of the way the diseases are often inherited and because mitochondria are so critical to cell function. The subclass of these diseases that have neuromuscular disease symptoms are often called a mitochondrial myopathy.
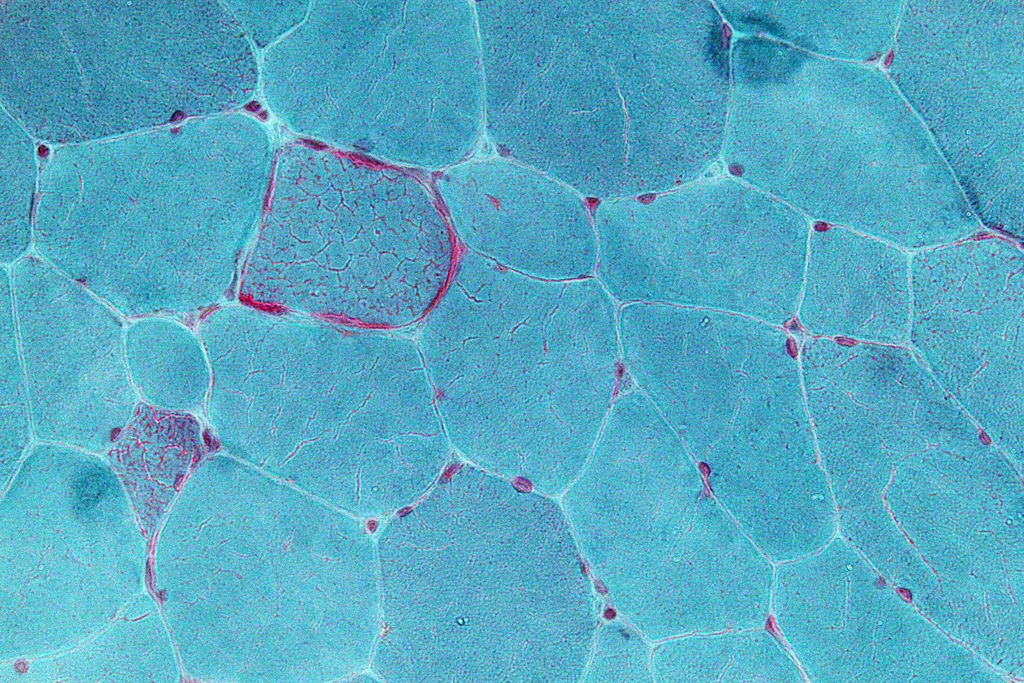
Image: Very high magnification micrograph showing ragged red fibers (also ragged red fibers), commonly abbreviated RRF, in a mitochondrial myopathy (Photo courtesy of Wikimedia Commons).
To address the lack of effective treatment for these disorders, investigators at the University of Cambridge (United Kingdom) exploited a recently developed mouse model (the m.5024C>T tRNAAla mouse) that recapitulates common molecular features of heteroplasmic mtDNA disease in cardiac tissue.
To modify the mitochondrial genome, the investigators chose a programmable nuclease therapy approach, using mitochondrially targeted zinc-finger nucleases (mtZFN), rather than the more frequently used CRISPR/Cas9 method. The mtZFN gene-editing tool was delivered to the mice via an adeno-associated virus vector. The tool was designed to recognize and then eliminate mutant mitochondrial DNA, based on sequence differences between healthy and mutant mitochondrial DNA.
The investigators reported in the September 24, 2018, online edition of the journal Nature Medicine that by systemically administering mitochondrially targeted mtZFN using adeno-associated virus to the mice, they successfully induced specific elimination of mutant mtDNA across the heart. The biochemical and physiological behavior of the diseased hearts returned to near normal values.
"One idea for treating these devastating diseases is to reduce the amount of mutated mitochondrial DNA by selectively destroying the mutated DNA, and allowing healthy DNA to take its place," said senior author Dr. Michal Minczuk, program leader at the mitochondrial biology unit of the University of Cambridge.
Related Links:
University of Cambridge