DNA Methylation Pattern May Be Biomarker for Cancer Detection
By LabMedica International staff writers
Posted on 17 Dec 2018
A team of Australian researchers has determined that a characteristic pattern of methylated DNA allows for easy detection of the genomes of cancerous cells and may serve as a universal biomarker for rapid diagnosis of many types of cancer.Posted on 17 Dec 2018
Methyl groups regulate gene expression and thereby control how a cell functions. Investigators at the University of Queensland (Brisbane, Australia) had reported previously that in healthy cells, methyl groups were spread out across the genome, but the genome of cancer cells were characterized by intense clusters of methyl groups at very specific locations. They named this distribution signature of the methylated DNA the "Methylscape."
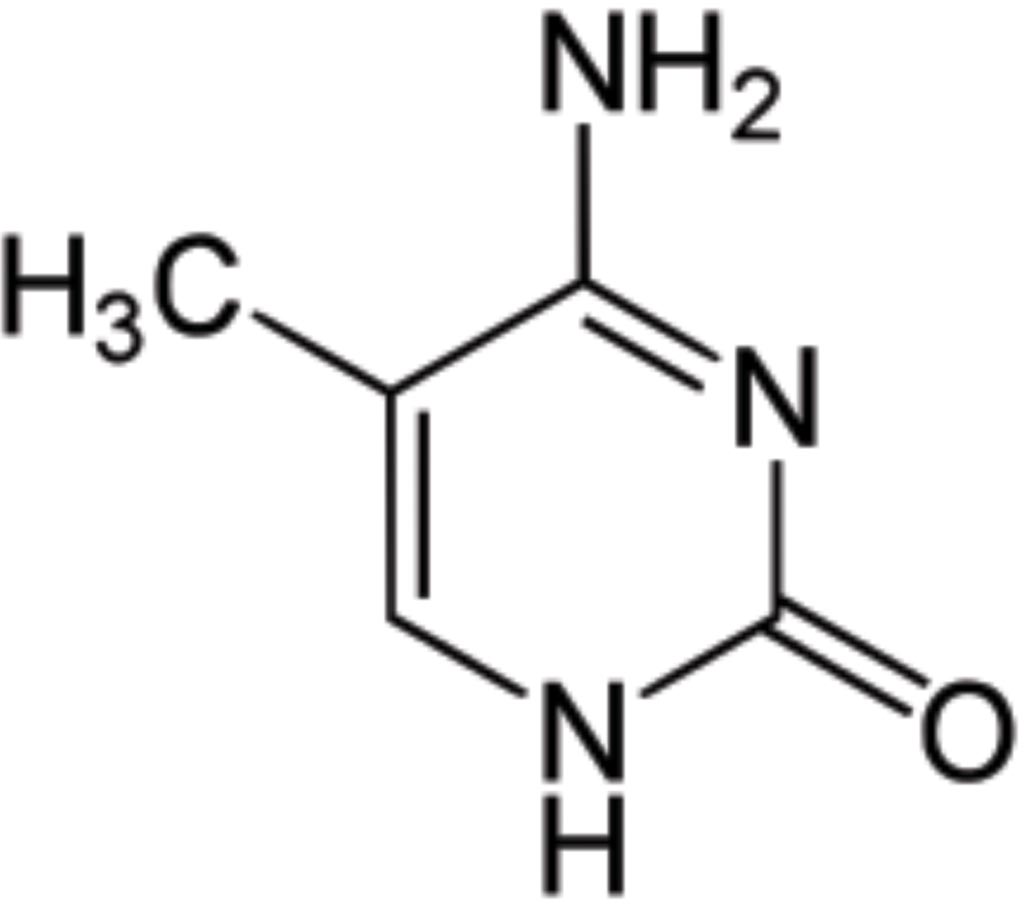
Image: 5-Methylcytosine is a methylated form of the DNA base cytosine that may be involved in the regulation of gene transcription (Photo courtesy of Wikimedia Commons).
The investigators examined the effect of levels and genomic distribution of methylcytosines on the physicochemical properties of DNA to detect the Methylscape biomarker. They found that DNA polymeric behavior was strongly affected by differential patterning of methylcytosine, leading to fundamental differences in DNA solvation and DNA-gold affinity between cancerous and normal genomes. One of the unique properties of the cancer cell Methylscape was its affinity for gold nanoparticles. Binding of DNA to such nanoparticles caused an immediate color change that was visible to the naked eye.
The investigators used electrochemical and colorimetric techniques to develop extremely simple, label-free, and naked eye platforms that could finely detect the Methylscape biomarker from cancer genomes based on the level of gold-DNA adsorption on planar and colloidal gold surfaces respectively. These assays were quick, i.e., analysis time less than 10 minutes, and required minimal sample preparation and small DNA input.
They tested these approaches on a large cohort of over 200 human samples (i.e., genomic DNAs extracted from cell-lines, tissues, and plasma) representing various cancer types. Results published in the December 4, 2018, online edition of the journal Nature Communications revealed that the accuracy of cancer detection was as high as 90%.
"Virtually every piece of cancerous DNA we examined had this highly predictable pattern. It seems to be a general feature for all cancer. It is a startling discovery," said senior author Dr. Matt Trau, professor of chemistry at the University of Queensland. "We certainly do not know yet whether it is the Holy Grail or not for all cancer diagnostics, but it looks really interesting as an incredibly simple universal marker of cancer, and as a very accessible and inexpensive technology that does not require complicated lab based equipment like DNA sequencing."
Related Links:
University of Queensland