Nanoscale Device Developed for Separation of NA Mixtures
By LabMedica International staff writers
Posted on 24 Mar 2017
A team of Japanese engineers has developed a nanoscale device for the rapid separation of microRNA (miRNA) from mixtures of miRNA, RNA, and DNA.Posted on 24 Mar 2017
MicroRNAs are a small noncoding family of 19- to 25-nucleotide RNAs that regulate gene expression by targeting messenger RNAs (mRNAs) in a sequence specific manner, inducing translational repression or mRNA degradation, depending on the degree of complementarity between miRNAs and their targets. Many miRNAs are conserved in sequence between distantly related organisms, suggesting that these molecules participate in essential processes. In fact, miRNAs have been shown to be involved in the regulation of gene expression during development, cell proliferation, apoptosis, glucose metabolism, stress resistance, and cancer.
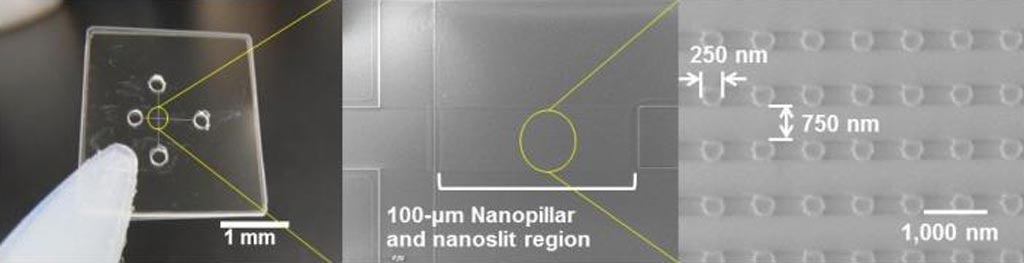
Image: Ultrafast electrophoretic microRNA extraction from a nucleic acids mixture using quartz nanopillars of 250-nanometer diameter arrayed inside a 100-nanommeter high nanoslit region (Photo courtesy of Dr. Noritada Kaji, Nagoya University).
Investigators at Nagoya University have developed an innovative nanoscale device that can rapidly separate microRNA from DNA/RNA mixtures obtained from cells.
The device was fabricated by electron beam lithography on a quartz substrate to contain a 25×100 micrometer array of "nanopillars" (small columns with a diameter of 250 nanometers and height of 100 nanometers) in shallow "nanoslits" with a height of 100 nanometers. This separation approach using a top-down fabrication technique enabled the precise control of DNA conformation during electrophoresis and demonstrated that the geometric pattern of the nanopillar array could control the separation mode and enhance the throughput. The square pattern improved the resolution of separation proportionally to the applied electric field and transferred the larger DNA molecules more rapidly than it transferred the smaller molecules of miRNA. Combining nanoslit structures provided an entropic trapping effect and improved the speed of separation and resolution.
Feasibility studies, using a mixture of total RNA and genomic DNA, were performed to elucidate whether this technique was applicable over a wide size range of nucleic acids. Results published in the March 8, 2017, online edition of the journal Scientific Reports revealed that a mixture of genomic DNA, total RNA, and miRNA from HeLa cells could be separated within 100 microseconds.
"We believe that the nanobiodevice separates microRNA from mixtures through a combination of two different physical behaviors of confined polymers in the nanopoillar array, non-equilibrium transport and entropic trapping," said contributing author Dr. Noritada Kaji, associate professor of engineering at Nagoya University. "The applied electric field combines with the unique nanostructure of the nanobiodevice to generate a strong electric force that induces rapid concentration and separation."