Screening Technique Finds Endothelial Cells Targeted by Nanoparticles
By LabMedica International staff writers
Posted on 18 Oct 2018
A recently developed technique allows researchers to identify lipid nanoparticles that preferentially target endothelial cells – rather than liver cells – for transport of the components of the CRISPR/Cas9 gene-editing tool.Posted on 18 Oct 2018
Nanoparticle-mediated delivery of siRNA to hepatocytes has been used to treat disease in humans. However, systemically delivering RNA drugs to tissues other than the liver has remained an important challenge, primarily because there is no high-throughput method to identify nanoparticles that deliver functional mRNA to cells in vivo.
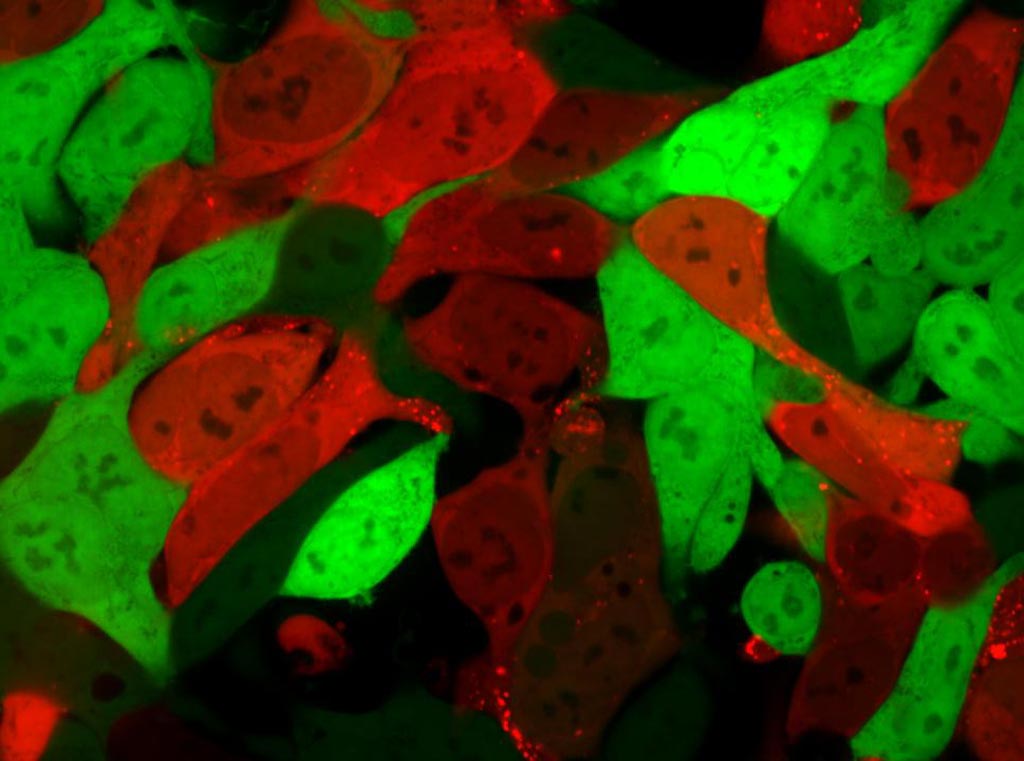
Image: Cells that are normally bright green turn bright red after lipid nanoparticles have delivered an mRNA cargo encoding Cre protein. Cells that are red contain the mRNA, while green cells do not (Photo courtesy of Daryll Vanover, Georgia Institute of Technology).
To increase the number of nanoparticles that could be studied in vivo, investigators at the Georgia Institute of Technology (Atlanta, USA) designed a high-throughput method for which they coined the name FIND (Fast Identification of Nanoparticle Delivery). This method is capable of quantifying how more than 100 lipid nanoparticles (LNPs) deliver mRNA that is translated into functional protein.
To prepare the nanoparticles nucleic acids (mRNA, DNA barcodes, siRNA, and sgRNA) were diluted in citrate buffer while lipid-amine compounds, alkyl-tailed PEG, cholesterol, and helper lipids were diluted in ethanol. The nanoparticles were generated by combining the citrate and ethanol phases by syringes in a microfluidic device.
The FIND technique also introduced a red-colored tracer protein (Cre) into cells. This was done by co-delivering Cas9 mRNA and single-guide RNA, which induced endothelial cell gene editing.
The investigators reported in the October 1, 2018, online edition of the journal Proceedings of the National Academy of Sciences of the United States of America that they had measured how more than 250 LNPs were able to deliver mRNA to multiple cell types in vivo and identified two LNPs that efficiently delivered siRNA, single-guide RNA (sgRNA), and mRNA to endothelial cells. One of the nanoparticles delivered Cas9 mRNA and sgRNA to splenic endothelial cells as efficiently as hepatocytes, distinguishing it from LNPs that delivered Cas9 mRNA and sgRNA to hepatocytes more than other cell types.
"We hope to take projects that would ordinarily require years and complete several of them within the next 12 months," said senior author Dr. James E. Dahlman, assistant professor of biomedical engineering at the Georgia Institute of Technology. "FIND could be used to carry all sorts of nucleic acid drugs into cells. That could include small RNAs, large RNAs, small DNAs and large DNAs - many different types of genetic drugs that are now being developed in research labs."
Related Links:
Georgia Institute of Technology