Mechanism Identified for Control of Nuclear Pore Complexes
By LabMedica International staff writers
Posted on 10 Oct 2018
Researchers have identified a molecular mechanism that helps control the number of nuclear pore complexes in a cell, a measure of some importance, since cells that transform into cancers often have an excess of these features.Posted on 10 Oct 2018
The total number of nuclear pore complexes (NPCs) per nucleus varies greatly between different cell types and is known to change during cell differentiation and cell transformation. However, the underlying mechanisms that control how many nuclear transport channels are assembled into a given nuclear envelope remain unclear.
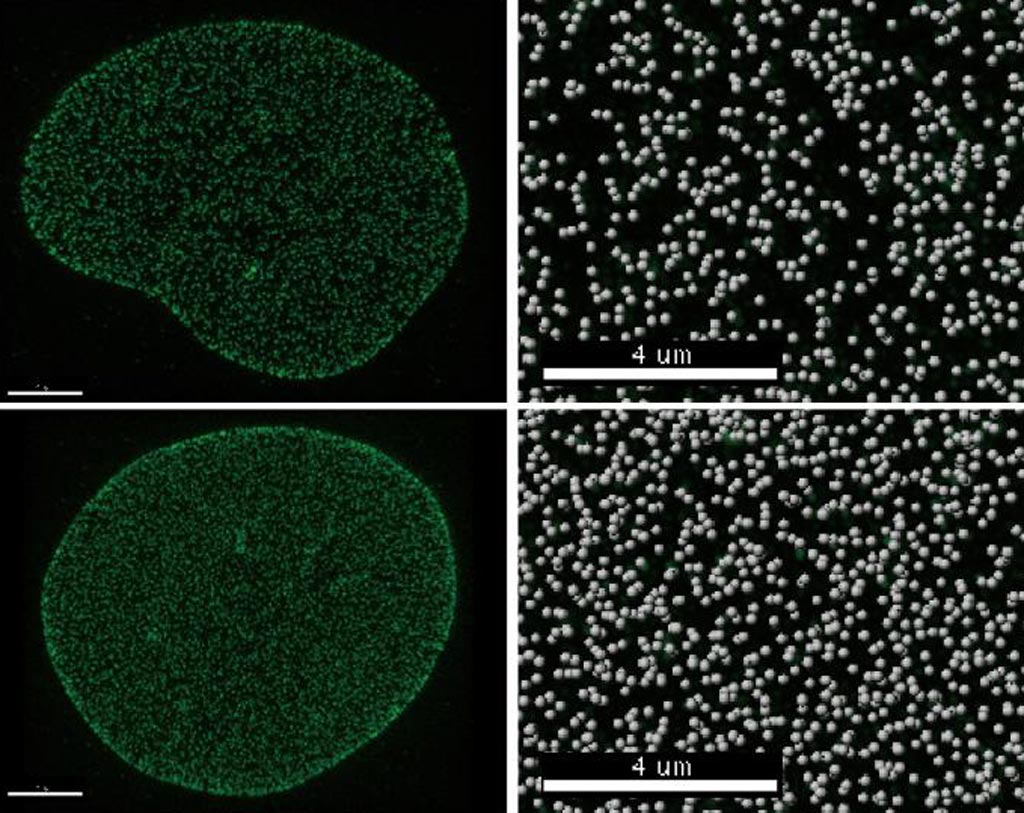
Image: Cells with the Tpr protein (top row) have fewer nuclear pores than cells without the protein (bottom row). The right column shows a close-up of the pore density, with many more pores appearing in the absence of Tpr (bottom left) (Photo courtesy of the Salk Institute for Biological Studies).
To study NPC control mechanisms, investigators at the Salk Institute for Biological Studies (La Jolla, CA, USA) focused their attention on the nucleoporin Tpr, which has been implicated in certain cancers. TPR is a component of the nuclear pore complex (NPC), a complex required for the trafficking across the nuclear envelope. Tpr functions as a scaffolding element in the nuclear phase of the NPC essential for normal nucleocytoplasmic transport of proteins and mRNAs, plays a role in the establishment of nuclear-peripheral chromatin compartmentalization in interphase, and in the mitotic spindle checkpoint signaling during mitosis. In association with the protein NUP153, Tpr is involved in the quality control and retention of unspliced mRNAs in the nucleus.
The investigators reported in the September 18, 2018, online edition of the journal Genes & Development that depletion of Tpr, but not Nup153, dramatically increased the total NPC number in various cell types. This negative regulation of Tpr occurred via a phosphorylation cascade of extracellular signal-regulated kinase (ERK), the central kinase of the mitogen-activated protein kinase (MAPK) pathway. Tpr served as a scaffold for ERK to phosphorylate the nucleoporin Nup153, which was critical for early stages of NPC biogenesis.
"Previously, we did not have the tools to artificially increase nuclear pores," said senior author Dr. Martin Hetzer, chief science officer at the Salk Institute for Biological Studies. "This is the first time that modifying a component within the transport channel has been shown to increase the number of nuclear pores. Our study provides an experimental avenue to ask critical questions: What are the consequences of boosting the number of nuclear pores in a healthy cell to mimic those found in a cancer cell? Does this affect gene activity? Why do cancer cells increase the number of nuclear pores?"
Related Links:
Salk Institute for Biological Studies