LncRNAs Linked to Prostate Cancer Progression
By LabMedica International staff writers
Posted on 06 Jun 2018
A long non-coding RNA has been linked to androgen receptor signaling and the growth and development of prostate cancer.Posted on 06 Jun 2018
Long non-coding RNAs (lncRNAs) are considered to be non-protein coding transcripts longer than 200 nucleotides. This somewhat arbitrary limit distinguishes lncRNAs from small regulatory RNAs such as microRNAs (miRNAs), short interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. LncRNAs have been found to be involved in numerous biological roles including imprinting, epigenetic gene regulation, cell cycle and apoptosis, and metastasis and prognosis in solid tumors. Most lncRNAs are expressed only in a few cells rather than whole tissues, or they are expressed at very low levels, making them difficult to study. Their name notwithstanding, long non-coding RNAs (lncRNAs) have been found to actually encode synthesis of small polypeptides that can fine-tune the activity of critical cellular components.
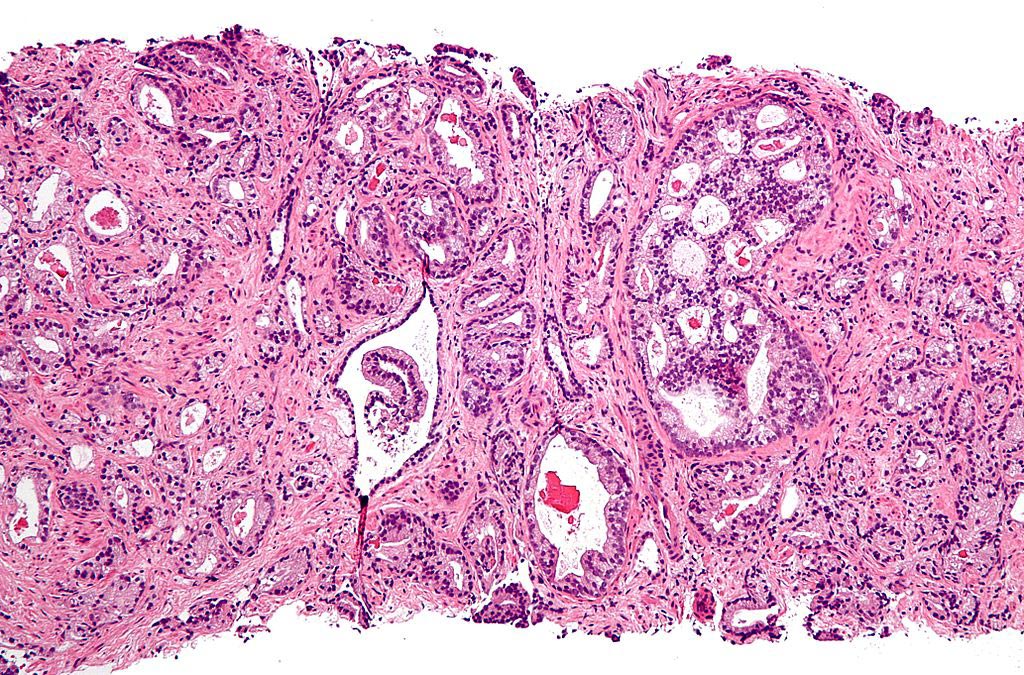
Image: A micrograph of prostate adenocarcinoma, acinar type, the most common type of prostate cancer (Photo courtesy of Wikimedia Commons).
The androgen receptor (AR) plays a critical role in the development of the normal prostate as well as prostate cancer. Investigators at the University of Michigan (Ann Arbor, USA) employed an integrative transcriptomic analysis of prostate cancer cell lines and tissues to identify the lncRNA ARLNC1 (AR-regulated long noncoding RNA 1) as being strongly associated with AR signaling in prostate cancer progression. Not only was ARLNC1 induced by the AR protein, but ARLNC1 stabilized the AR transcript via RNA–RNA interaction.
The investigators further reported in the May 28, 2018, online edition of the journal Nature Genetics that in mouse models, overexpression of ARLNC1 prompted formation of large tumors, while knockdown of this lncRNA suppressed AR expression, global AR signaling, and prostate cancer growth. These findings supported a role for ARLNC1 in potentiating AR signaling during prostate cancer progression and identified ARLNC1 as a novel therapeutic target.
"The androgen receptor is an important target in prostate cancer. Understanding that target is important. This study identifies a feedback loop that we could potentially disrupt as an alternative to blocking the androgen receptor directly," said senior author Dr. Arul Chinnaiyan, professor of pathology and urology at the University of Michigan. "At the end of the day, you are creating or stabilizing more androgen receptor signaling in general and driving this oncogenic pathway forward. We are envisioning a potential therapy against ARLNC1 in combination with therapy to block the androgen receptor - which would hit the target and also this positive feedback loop. There are a number of these lncRNAs that we do not understand how they functionally work. Some of them will certainly be very useful as cancer biomarkers and we think a subset is important in biological processes."
Related Links:
University of Michigan