Enzyme Structure May Lead to Control of DNA Methylation Process
By LabMedica International staff writers
Posted on 21 Feb 2018
The recently established molecular structure of the enzyme DNA methyltransferase 3A bound to its DNA substrate is expected to eventually lead to the ability to control DNA methylation content and, by extension, gene expression and cell differentiation.Posted on 21 Feb 2018
DNA methylation catalyzed by the de novo DNA methyltransferases 3A (DNMT3A) and 3B (DNMT3B) at cytosine residues is essential for genome regulation and development. Disruption of this process has been implicated in various diseases, notably cancer. However, the mechanisms underlying DNMT3 substrate recognition and enzymatic specificity have remained elusive.
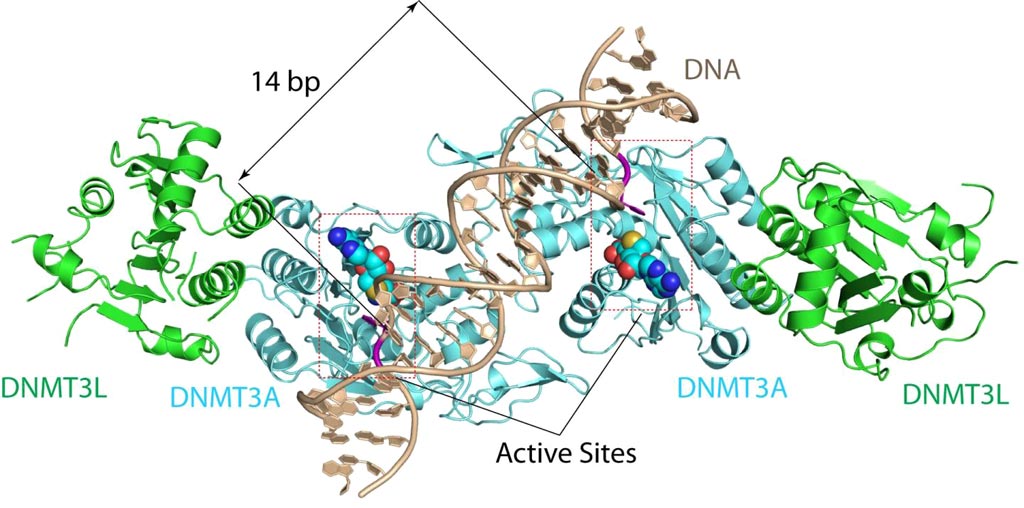
Image: The DNMT3A-DNA complex. The structure reveals that DNMT3A molecules attack two substrate sites adjacent to each other on the same DNA molecule. DNMT3L (green) is a regulatory protein of DNMT3A (Photo courtesy of the Song Laboratory, University of California, Riverside).
To better understand this mechanism, investigators at the University of California, Riverside (USA) solved the 2.65-ångström crystal structure of the DNMT3A–DNMT3L–DNA complex. They described in the February 7, 2018, online edition of the journal Nature the state in which two DNMT3A monomers simultaneously attacked two cytosine–phosphate–guanine (CpG) dinucleotides, with the target sites separated by 14 base pairs within the same DNA duplex.
The DNMT3A–DNA interaction involved a target recognition domain, a catalytic loop, and DNMT3A homodimeric interface. A specific arginine residue in the target recognition domain was shown to make crucial contacts with CpG, ensuring DNMT3A's enzymatic preference towards CpG sites in cells.
"The structure reveals that DNMT3A molecules attack two substrate sites adjacent to each other on the same DNA molecule," said senior author Dr. Jikui Song, associate professor of biochemistry at the University of California, Riverside. "This now offers us a much clearer view on how de novo DNA methylation takes place. Our work presents the first structural view of de novo DNA methylation and presents a model for how some DNMT3A mutations contribute to cancers, such as acute myeloid leukemia. This study should provide important insights into the function of DNMT3B as well."
"Before our study, why mammalian DNA methylation mostly occurs at the CpG sites was not understood, and our understanding of de novo DNA methylation was purely based on computational modeling, which cannot reliably explain how DNMT3A works," said Dr. Song. "Just how DNMT3A succeeded in binding to its substrate was not understood either. Our structure for DNMT3A-DNA complex addresses all these concerns, offering a far better understanding of how specific DNA methylation patterns are generated."
Related Links:
University of California, Riverside