Computer Program Confirms Relationship of cfDNA and Tumor DNA
By LabMedica International staff writers
Posted on 16 Nov 2017
Cancer researchers have developed an accurate, scalable approach for monitoring cell-free tumor DNA from blood samples.Posted on 16 Nov 2017
Development of this approach was based on the hypothesis that whole-exome sequencing of cell-free DNA (cfDNA) could enable comprehensive profiling of tumors from blood. However, the genome-wide concordance between cfDNA in blood samples and tumor biopsies was uncertain.
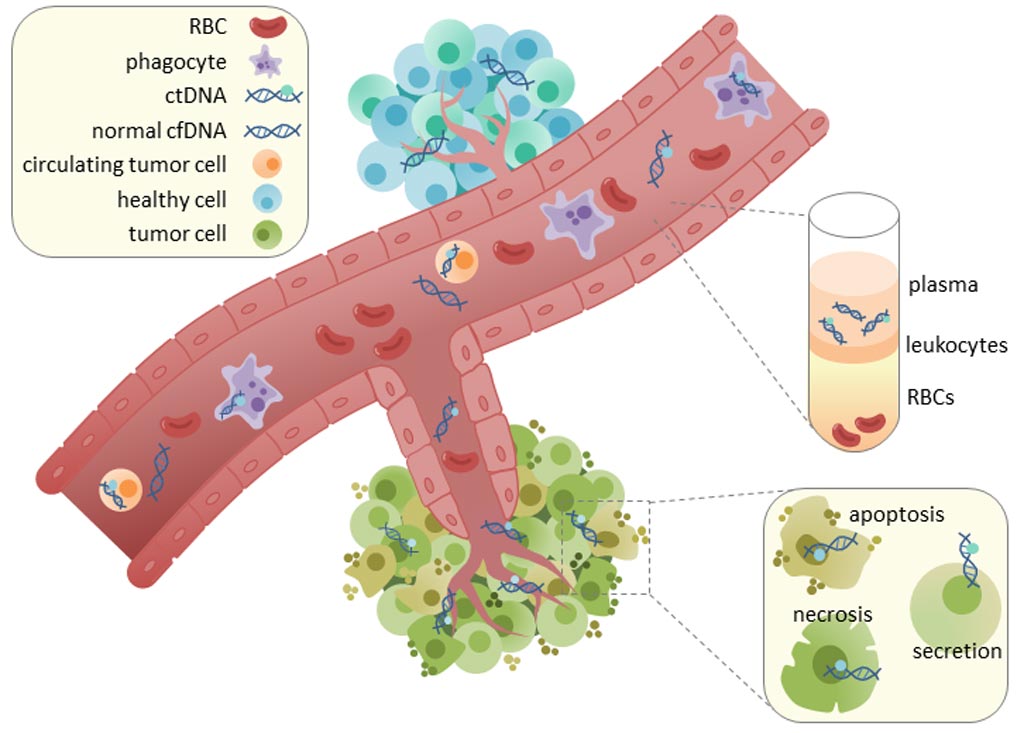
Image: Circulating tumor DNA (ct or cfDNA) is found in serum and plasma fractions from blood. The mechanism of cfDNA release is unknown, though apoptosis, necrosis, and active secretion from tumor cells have been hypothesized (Photo courtesy of Wikimedia Commons).
To confirm the relationship between cfDNA in the blood and DNA from cancer cells in tumors, investigators at the Broad Institute of the Massachusetts Institute of Technology and Harvard University (Cambridge, MA, USA) developed ichorCNA, a computer program able to analyze DNA fragments for mutation patterns nearly universal in cancer genomes, and as a result able to identify cancers with both known and unknown mutations. The ichorCNA, software quantified tumor content in cfDNA from 0.1× coverage whole-genome sequencing data without prior knowledge of tumor mutations.
The investigators reported in the November 6, 2017, online edition of the journal Nature Communications, that they used ichorCNA to analyze 1439 blood samples from 520 patients with metastatic prostate or breast cancers. In the earliest tested sample for each patient, 34% of patients had at least 10% tumor-derived cfDNA, sufficient for standard coverage whole-exome sequencing.
Using whole-exome sequencing, the investigators validated the concordance of clonal somatic mutations (88%), copy number alterations (80%), mutational signatures, and neoantigens between cfDNA and matched tumor biopsies from 41 patients that had more than 10% tumor cfDNA in the serum.
"Our study has demonstrated that we can get a cancer whole exome reliably, from blood; that it reflects the matched tumor biopsy; and that it can be done for a significant fraction of patients with metastatic cancer," said first author Dr. Viktor Adalsteinsson, leader of the blood biopsy team at the Broad Institute. "This validation suggests that we can use blood biopsies for large-scale genomic characterization of disease in patients with metastatic cancer. Our ultimate hope is to use blood biopsies to exhaustively search for and characterize even the smallest remnants of tumors, and, as tumors evolve in more advanced stages of cancer, developing resistance or becoming metastatic, we might access time points that could be pivotal in deciding which therapies are right for that patient."
Related Links:
Broad Institute