Two Critical Genes Identified for Soft Tissue Infections
By LabMedica International staff writers
Posted on 04 Oct 2017
A team of molecular microbiologists has identified two genes that are critical for infection of soft tissue and dissemination into the bloodstream by pathogenic Group A Streptococcus bacteria.Posted on 04 Oct 2017
The World Health Organization ranks the Group A Streptococcus (GAS) in the top 10 leading causes of morbidity and mortality from infectious diseases worldwide. GAS is a strict human pathogen causing both benign superficial infections as well as life-threatening invasive diseases. All GAS infections begin by colonization of an epithelium (throat or skin) followed by propagation into subepithelial tissues.
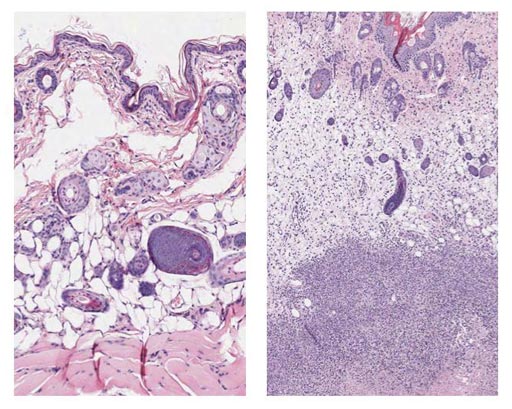
Image: Tissue staining shows Group A Streptococcus soft tissue infection at the cellular level. (L-R) Uninfected mouse tissue and mouse tissue 48 hours after infection. The dense dots indicate immune system cells that swarmed in to attempt to control the infection. The densest purple staining toward the bottom is necrotic tissue surrounding bacteria (Photo courtesy of Joshua Leiberman, University of Maryland).
Investigators at the University of Maryland (College Park, USA) used transposon-sequencing (Tn-seq) to define the genetic requirements for in vivo fitness of GAS in subepithelial tissue.
Transposon sequencing begins by transforming bacterial populations with transposable elements using bacteriophages. The technique requires the creation of a transposon insertion library, which will contain a group of mutants that collectively have transposon insertions in all non-essential genes. The library is grown under the condition that is of interest. Mutants with transposons inserted in genes required for growth under the test condition will diminish in frequency from the population. To identify genes being lost, sequences encompassing the transposon ends are amplified by polymerase chain reaction (PCR) and sequenced by massive parallel sequencing (MPS) to determine the location and abundance of each insertion mutation. The importance of each gene for growth under the test condition is determined by comparing the abundance of each mutant before and after growth under the condition being examined.
The investigators reported in the August 23, 2017, online edition of the journal PLOS Pathogens that they had identified 273 subcutaneous fitness (scf) genes with 108 of those previously of “unknown function”. Two yet uncharacterized genes, scfA and scfB, were shown to be critical during soft tissue infection and dissemination into the bloodstream. The investigators hypothesized that that these scfAB genes play an integral role in enhancing adaptation and fitness of GAS during localized skin infection, and potentially in propagation to other deeper host environments.
"Invasion under the skin, or subcutaneously, is not the norm for Group A Streptococcus bacteria; it is actually very rare," said senior author Dr. Kevin McIver, professor of cell biology and molecular genetics at the University of Maryland. "We hypothesized that there must be genes in the bacteria important for invading soft tissues and surviving under the skin. And we tested that theory by using transposons to make thousands of different individual mutants that we used to infect a subcutaneous environment in mice. Transposon sequencing can be used to probe how bacteria infect humans in any environment you can think of. Like Group A Streptococcus, many pathogenic bacteria have completely sequenced genomes, but we do not know what most of the genes are doing. We are excited to have a method to interrogate all that unknown genetic material to better understand human infections."
Related Links:
University of Maryland