Fruit Fly Model Used to Detect Proteins Coded by Circular RNAs
By LabMedica International staff writers
Posted on 06 Apr 2017
An international team of molecular biologists used a Drosophila (fruit fly) model to demonstrate that proteins could be synthesized from genetic information contained in molecules of circular RNA (circRNA).Posted on 06 Apr 2017
CircRNAs in animals form a large class of particularly stable RNAs produced by circularization of specific exons. Typically, circRNAs arise from otherwise protein-coding genes, but circular RNAs produced in the cell had not been shown previously to code for proteins and were categorized as noncoding RNAs. While some circular RNAs have recently shown potential as gene regulators the biological function of most circular RNAs was unclear.
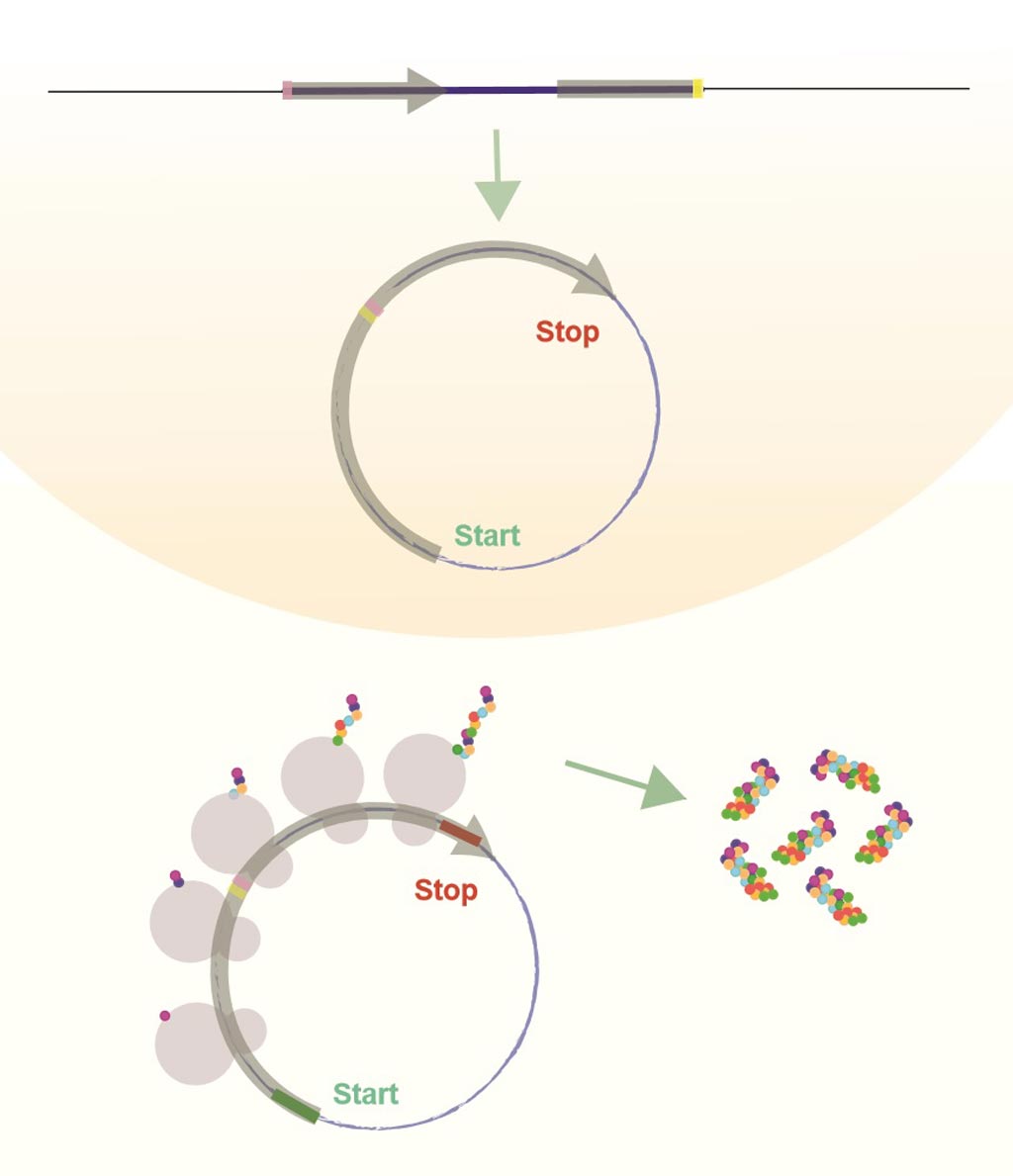
Image: Circular RNAs (circRNA) are produced in the cell nucleus after they are copied from the DNA and closed. Some circRNAs are translated and produce protein once they are exported from the nucleus (Photo courtesy of Dr. Sebastian Kadener, Hebrew University of Jerusalem).
Investigators at the Hebrew University of Jerusalem and their colleagues at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association used a fruit fly model system and developed or adapted various techniques from molecular biology, computational biochemistry, and neurobiology to show that specific circRNA molecules were bound to ribosomes, the cellular organelles responsible for protein synthesis.
Furthermore, the investigators detected proteins produced from these molecules. Since circular RNAs do not have 5' or 3' ends, they are resistant to exonuclease-mediated degradation and are presumably more stable than most linear RNAs in cells.
In particular, the investigators reported in the March 23, 2017, online edition of the journal Molecular Cell that a circRNA generated from the muscleblind locus encoded a protein, which they detected in fly head extracts by mass spectrometry.
Senior author Dr. Sebastian Kadener, professor of biological chemistry at the Hebrew University of Jerusalem, said, "By identifying the function of circRNAs, this research helps advance our understanding of molecular biology, and can be helpful in understanding aging or neurodegenerative diseases."
Dr. Nikolaus Rajewsky, professor for systems biology at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association said, "We think that translation of circRNAs is very interesting and that its prevalence and importance must be further investigated."