Antibiotic Characterized for Drug-Resistant Strains of TB
By LabMedica International staff writers
Posted on 01 Oct 2018
An international team of biochemists and molecular biologists has isolated and characterized a novel, naturally occurring antibiotic that is capable of killing multidrug-resistant strains of Mycobacterium tuberculosis.Posted on 01 Oct 2018
Antibiotic-resistant bacterial pathogens pose an urgent healthcare threat, prompting a demand for new medicines. In particular, development of rifampicin resistance in M. tuberculosis has complicated treatment of this disease, since it extends treatment time for tuberculosis from six months to two years.
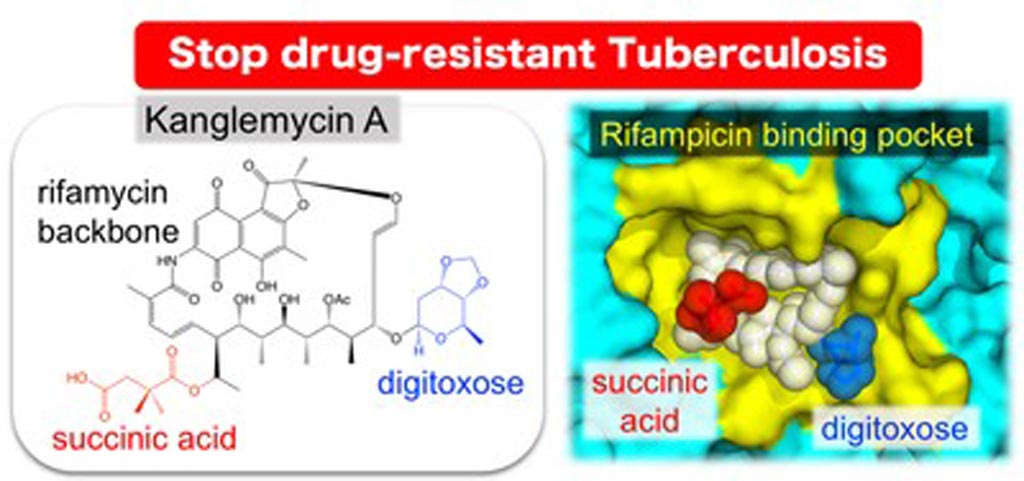
Image: The natural antibiotic kanglemycin A binds bacterial RNA polymerase at the rifampicin binding-pocket, but maintains potency against rifampicin-resistant mutants due to two unique chemical groups (digitoxose and succinic acid) that increase its affinity to rifampicin-resistant RNA polymerase by binding just outside the rifampicin-binding pocket (Photo courtesy of the Murakami Laboratory, Pennsylvania State University).
In searching for new drug candidates to treat tuberculosis, investigators screened a library of naturally occurring compounds from the biotechnology company Demuris Ltd. (Newcastle upon Tyne, United Kingdom) for their ability to inhibit bacterial cell growth or prevent the production of RNA.
The investigators reported in the September 20, 2018, online edition of the journal Molecular Cell that by using X-ray crystallography as well as biochemical and molecular biological techniques, they had identified the antibiotic kanglemycin A (KglA) as being effective against Mycobacterium tuberculosis. KglA, which is related to rifampicin, demonstrated antibiotic activity against rifampicin-resistant Gram-positive bacteria and multidrug-resistant (MDR-M) tuberculosis).
Rifamycins are a subclass of ansamycins with high potency against mycobacteria. This resulted in their widespread use in the treatment of tuberculosis, leprosy, and AIDS-related mycobacterial infections. Ansamycins were named for their unique structure, which consists of an aromatic moiety bridged by an aliphatic chain. The main difference between various derivatives of ansamycins is the aromatic moiety, which can be a naphthalene ring or a naphthoquinone ring as in rifamycin and the naphthomycins. The rifamycin group includes the "classic" rifamycin drugs as well as the rifamycin derivatives rifampicin (or rifampin), rifabutin, rifapentine, rifalazil, and rifaximin.
X-ray crystallography was used to determine the three-dimensional structure of the complex of kanglemycin A bound to a bacterial RNA polymerase enzyme. The X-ray crystal structures of KglA with the Escherichia coli RNA polymerase holoenzyme and Thermus thermophilus RNA polymerase-promoter complex revealed an altered -compared with rifampicin - conformation of KglA within the rifampicin-binding pocket.
"The X-ray structure actually revealed that kanglemycin A has two modifications that improve its function compared to rifampicin," said contributing author Dr. Katsuhiko Murakami, professor of biochemistry and molecular biology at Pennsylvania State University (University Station, USA). "First, one of modifications allows it to bind just outside of the rifampicin binding pocket increasing the strength of its affinity to the RNA polymerase in rifampicin-resistant bacteria. Second, another modification actually allows kanglemycin A to stop the synthesis of RNA even earlier than rifampicin. Understanding how kanglemycin A manages to maintain its affinity to rifampicin-resistant RNA polymerase and stay active against the drug-resistant bacteria will help to accelerate its approval for use in patients with tuberculosis."
"It is a really exciting finding," said senior author Dr. Nikolay Zenkin, professor of molecular biology at Newcastle University (United Kingdom). "The previously unknown interactions of the unique chemical groups of kanglemycin A with RNA polymerase will direct the development of antibiotics against rifampicin-resistant M. tuberculosis. Approximately one third of the world's population is already infected with M. tuberculosis, and 600,000 people every year are diagnosed with rifampicin-resistant tuberculosis. Our work is the first step in developing a new drug for the treatment of these patients."
The investigators expressed the belief that KglA represents a key starting point for the development of a new class of ansa-chain derivatized ansamycins to deal with the problem of rifampicin resistance.
Related Links:
Demuris
Pennsylvania State University
Newcastle University